Our research group uses genomic approaches to address research questions around three main themes; 1) evolutionary history, phylogenomics and macroevolution of species and evolutionary lineages, 2) genome evolution and the genomic basis of evolutionary processes, and 3) conservation genomics, genetic health and genetic management of threatened species. We work across a variety of animal groups, including extinct and living populations of mammals, birds, reptiles, and more. Below are some examples of major ongoing projects in the lab.
Reconstructing historical extinction using museum genomics
Australia has the highest rate of historical mammal extinction in the world. As part of various ongoing projects, our research uses museum genomics to reconstruct the evolution, demographic history, genetic health and decline of extinct Australian mammals, to understand diversity lost since European invasion of Australia in 1788.
E Roycroft et al. (2021) Museum genomics reveals the rapid decline and extinction of Australian rodents since European settlement. PNAS. https://doi.org/10.1073/pnas.2021390118
E Roycroft et al. (2022) Sequence capture from historical museum specimens: maximizing value for population and phylogenomic studies. Front Ecol Evol https://doi.org/10.3389/fevo.2022.931644
This work is supported by ARC Linkage Project LP180100315, ARC DECRA project DE240100573, and BioPlatforms Australia (as part of the Oz Mammals Genomics Initiative)

Genomic impact of island isolation
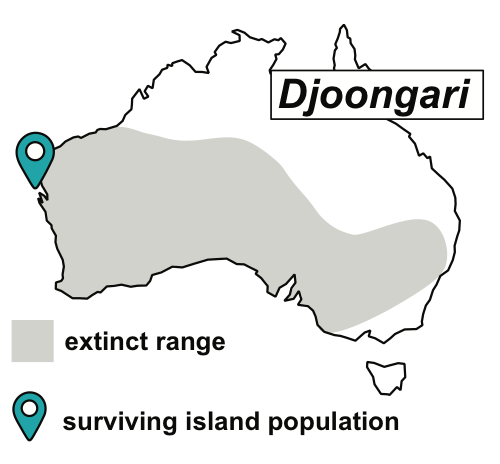
Australian offshore islands, largely free from introduced predators, protect many threatened species from extinction. Isolated from their mainland relatives for at least 8,000 years by rising sea levels, islands have been natural ‘arks’ for species that are now extinct or have severely declined on mainland Australia. These ‘island arks’ now have immense conservation significance, but little is known about their demographic history, genetic diversity and burden of deleterious mutations. As part of several ongoing projects, we are investigating the genome-wide impact of island isolation and small population size in a number of threatened Australian mammal and bird species.
E Roycroft et al. (Submitted) Remnant island populations purge genetic load.
This work is supported by ARC Linkage Project LP180100315, ARC DECRA project DE240100573, and the L’Oréal–UNESCO For Women in Science Fellowship.
Phylogenomics and diversification of Australian rodents

The old endemic rodents of Sahul (Australia and New Guinea) are approximately 160 species in 37 genera, and are the result of a single over-water colonisation event from southeast Asia ~ 7-9 million years ago. Rapid accumulation of lineages following initial colonisation of Sahul, and subsequent colonisations of present-day mainland Australia and nearby islands, have made phylogenetic relationships and biogeographic history of this group particularly difficult to resolve. As part of this ongoing project, we have generated species-level data from across all ~160 described species and many cryptic lineages in the Hydromyini to investigate diversification, molecular evolution and biogeography of Australian rodents.
E Roycroft et al. (2022) New Guinea uplift opens ecological opportunity across a continent. Current Biology. https://doi.org/10.1016/j.cub.2022.08.021
E Roycroft et al. (2020) Phylogenomics uncovers confidence and conflict in the rapid radiation of Australo-Papuan rodents. Systematic Biology. https://doi.org/10.1093/sysbio/syz044
These projects are funded by BioPlatforms Australia (as part of the Oz Mammals Genomics Initiative).
Diversification and extinction in the iconic Australian marsupials
Collaborators: Mark Eldridge (Australian Museum), Sally Potter (Macquarie University), Craig Moritz (ANU), Simon Ho (University of Sydney), Matthew Phillips (QUT) and the Oz Mammals Genomics Consortium
We are investigating diversification, biogeography and extinction at a continental-scale with species-level sampling of the iconic radiation of Australian marsupials.
More details soon…
This project is funded by BioPlatforms Australia (as part of the Oz Mammals Genomics Initiative).
Photo: Australian Museum, Petrogale xanthopus

Comparative genomics and chromosome evolution in Gehyra geckos
Collaborators: Craig Moritz (ANU), Janine Deakin (University of Canberra), and the Australian Reptile and Amphibian Genomics (AusARG) Consortium

The genus Gehyra represents a major radiation among Australian lizards and a subject of studies in chromosome change, speciation, adaptive evolution and sex chromosome evolution for 40 years. Recent phylogenomic and taxonomic studies have resolved species boundaries and relationships among the 62 lineages (49 described species).
We are generating high-quality reference genomes and whole-genome resequencing from across the Gehyra radiation, to investigate genome evolution, chromosome rearrangements, introgression, historical demography and predictors of genetic diversity.
This project is funded by BioPlatforms Australia via the AusARG initiative, the Australian Research Council (Discovery Project DP190102395) and by an Ignition Grant from the Centre for Biodiversity Analysis.
Photo: Sam Gordon, Gehyra purpurascens
Phylogeography and conservation genomics of Australian rodents
Collaborators: Fred Ford (NSW Department of Planning and Environment), Phoebe Burns (Zoos Victoria), Marissa Parrott (Zoos Victoria), Bill Breed (University of Adelaide), Kevin Rowe (Museums Victoria), Craig Moritz (Australian National University), in collaboration with the National Pookila Recovery Team.
Phylogeographic studies can provide powerful insight into how environment drives speciation across climatic contexts, and Australia is a model system for reconstructing the impact of past and present environments on diversification. We are investigating how fluctuating climates during the Pleistocene in Australia has contributed to speciation and demographic history in Australian rodents, especially in the delicate mouse group (Pseudomys delicatulus, P. novaeholladiae, P. hermannsbergensis, P. bolami), pebble-mound mice, chestnut mice (P. gracilicaudatus and P. nanus) and rock-rats (Zyzomys spp).
We have a special focus on conservation genomics of the threatened Australian pookila (New Holland mouse, P. novaehollandiae) across its range, especially in Victoria – where the species has seen dramatic declines in recent years. Using approaches integrating museum specimens, contemporary tissues and biobanked samples, we are investigating the past and ongoing loss of diversity in this species to inform the ongoing Pookila conservation breeding and genetic rescue program at Zoos Victoria, in collaboration with the National Pookila Recovery Team.
PA Burns, KC Rowe, ML Parrott, E Roycroft (2023) Population genomics of decline and local extinction in the endangered Australian Pookila. Biological Conservation 284, 110183. https://doi.org/10.1016/j.biocon.2023.110183
These projects are funded by the Native Australian Animals Trust, Holsworth Wildlife Research Endowment, Zoos Victoria, and BioPlatforms Australia (as part of the Oz Mammals Genomics Initiative).
Photo: Phoebe Burns, a pookila (Pseudomys novaehollandiae) during population monitoring in Victoria.

Past projects
Genomics of adaptive evolution and convergence in murine rodents

Adaptive radiations are characterised by the diversification and ecological differentiation of species, and replicated cases of this process provide natural experiments for understanding the repeatability and pace of molecular evolution. During adaptive radiation, genes related to ecological specialisation may be subject to recurrent positive directional selection. However, it is not clear to what extent patterns of lineage-specific ecological specialisation (including phenotypic convergence) are correlated with shared signatures of molecular evolution.
Using whole exome sequencing, we examined how molecular evolution is correlated with diversification in the spectacular adaptive radiation of murine rodents.
E Roycroft et al. (2021) Molecular Evolution of Ecological Specialisation: Genomic Insights from the Diversification of Murine Rodents. Genome Biology and Evolution. https://doi.org/10.1093/gbe/evab103
EEK Kopania et al. (preprint) Molecular evolution of male reproduction across species with highly divergent sperm morphology in diverse murine rodents. biorXiv. https://www.biorxiv.org/content/10.1101/2023.08.30.555585
This project was funded by NSF grant DEB-1754096 and DEB-1754393: ‘Rates of lineage, phenotypic, and genomic diversification in replicated radiations of murine rodents’.

